mirror of
https://github.com/datahub-project/datahub.git
synced 2025-12-25 08:58:26 +00:00
feat(ingest): dbt - adding support for dbt tests (#5201)
This commit is contained in:
parent
e2f1da00ff
commit
0ee2569d5c
@ -8,7 +8,9 @@ module.exports = {
|
||||
favicon: "img/favicon.ico",
|
||||
organizationName: "linkedin", // Usually your GitHub org/user name.
|
||||
projectName: "datahub", // Usually your repo name.
|
||||
stylesheets: ["https://fonts.googleapis.com/css2?family=Manrope:wght@400;600&display=swap"],
|
||||
stylesheets: [
|
||||
"https://fonts.googleapis.com/css2?family=Manrope:wght@400;600&display=swap",
|
||||
],
|
||||
themeConfig: {
|
||||
colorMode: {
|
||||
switchConfig: {
|
||||
@ -198,7 +200,10 @@ module.exports = {
|
||||
],
|
||||
],
|
||||
plugins: [
|
||||
["@docusaurus/plugin-ideal-image", { quality: 100, sizes: [320, 640, 1280, 1440, 1600] }],
|
||||
[
|
||||
"@docusaurus/plugin-ideal-image",
|
||||
{ quality: 100, sizes: [320, 640, 1280, 1440, 1600] },
|
||||
],
|
||||
"docusaurus-plugin-sass",
|
||||
[
|
||||
"docusaurus-graphql-plugin",
|
||||
|
||||
@ -39,4 +39,26 @@ We support the below actions -
|
||||
|
||||
Note:
|
||||
1. Currently, dbt meta mapping is only supported for meta configs defined at the top most level or a node in manifest file. If that is not preset we will look for meta in the config section of the node.
|
||||
2. For string based meta properties we support regex matching.
|
||||
2. For string based meta properties we support regex matching.
|
||||
|
||||
### Integrating with dbt test
|
||||
|
||||
To integrate with dbt tests, the `dbt` source needs access to the `run_results.json` file generated after a `dbt test` execution. Typically, this is written to the `target` directory. A common pattern you can follow is:
|
||||
1. Run `dbt docs generate` and upload `manifest.json` and `catalog.json` to a location accessible to the `dbt` source (e.g. s3 or local file system)
|
||||
2. Run `dbt test` and upload `run_results.json` to a location accessible to the `dbt` source (e.g. s3 or local file system)
|
||||
3. Run `datahub ingest -c dbt_recipe.dhub.yaml` with the following config parameters specified
|
||||
* test_results_path: pointing to the run_results.json file that you just created
|
||||
|
||||
The connector will produce the following things:
|
||||
- Assertion definitions that are attached to the dataset (or datasets)
|
||||
- Results from running the tests attached to the timeline of the dataset
|
||||
|
||||
#### View of dbt tests for a dataset
|
||||
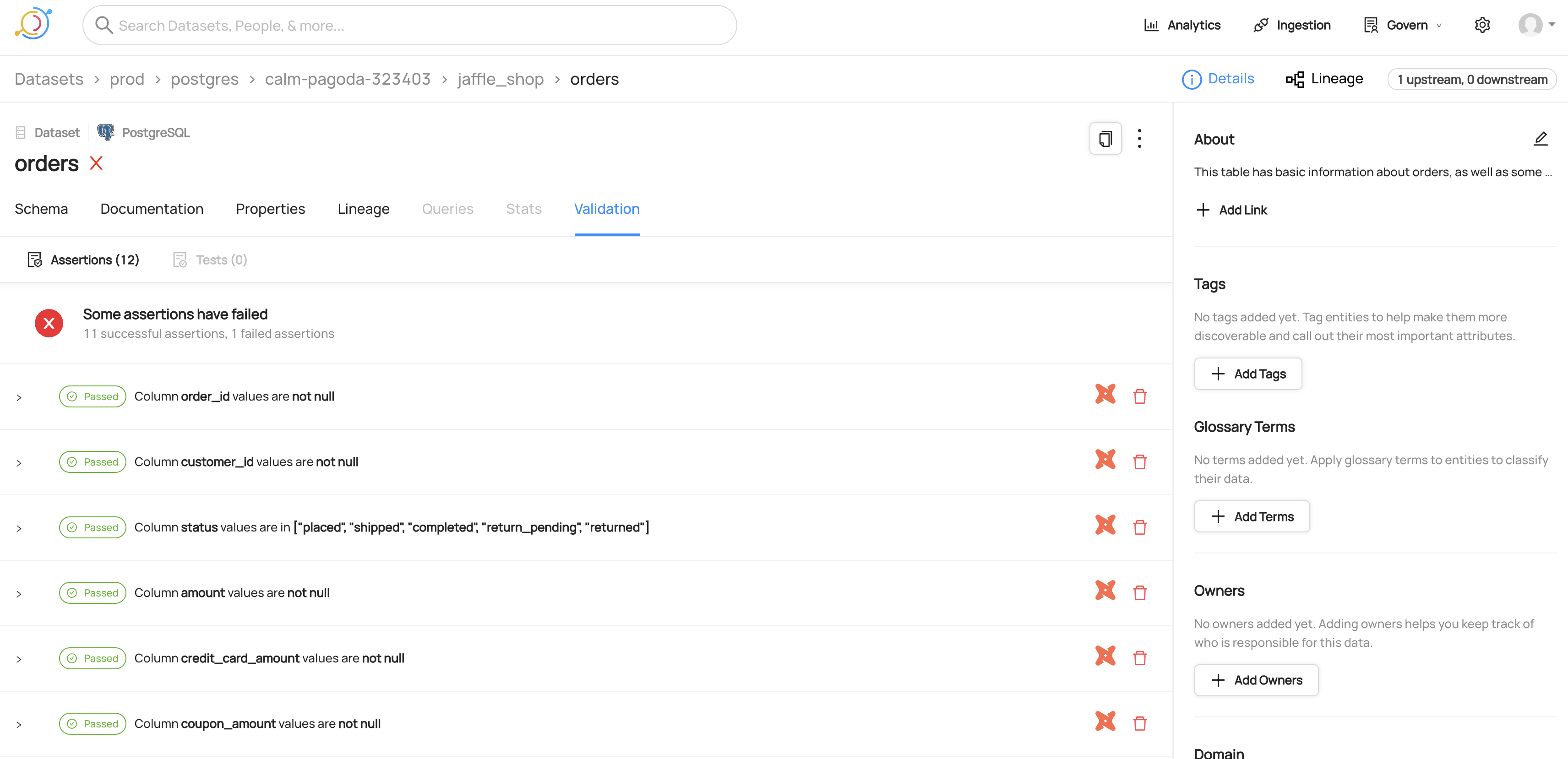
|
||||
#### Viewing the SQL for a dbt test
|
||||
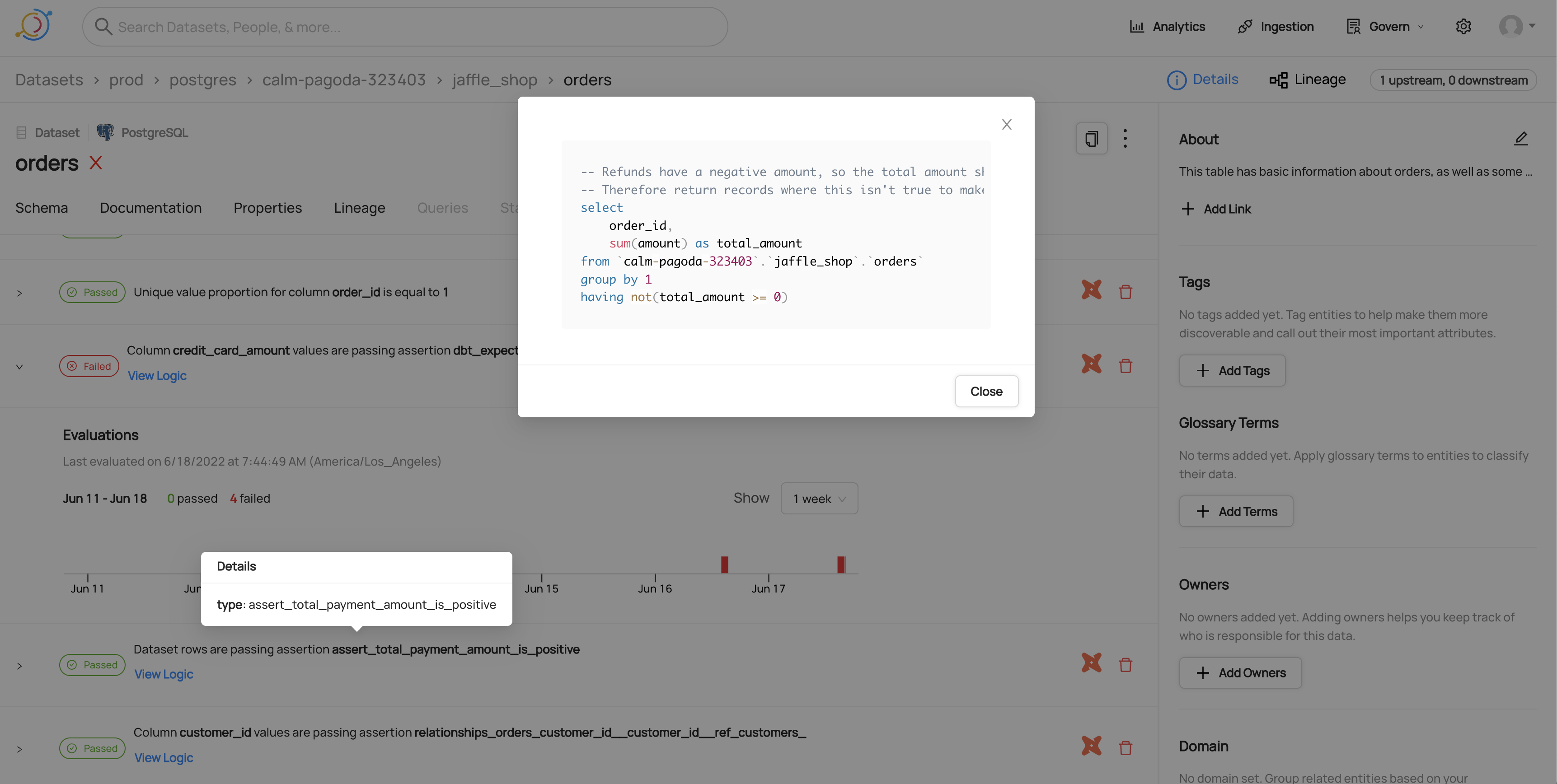
|
||||
#### Viewing timeline for a failed dbt test
|
||||
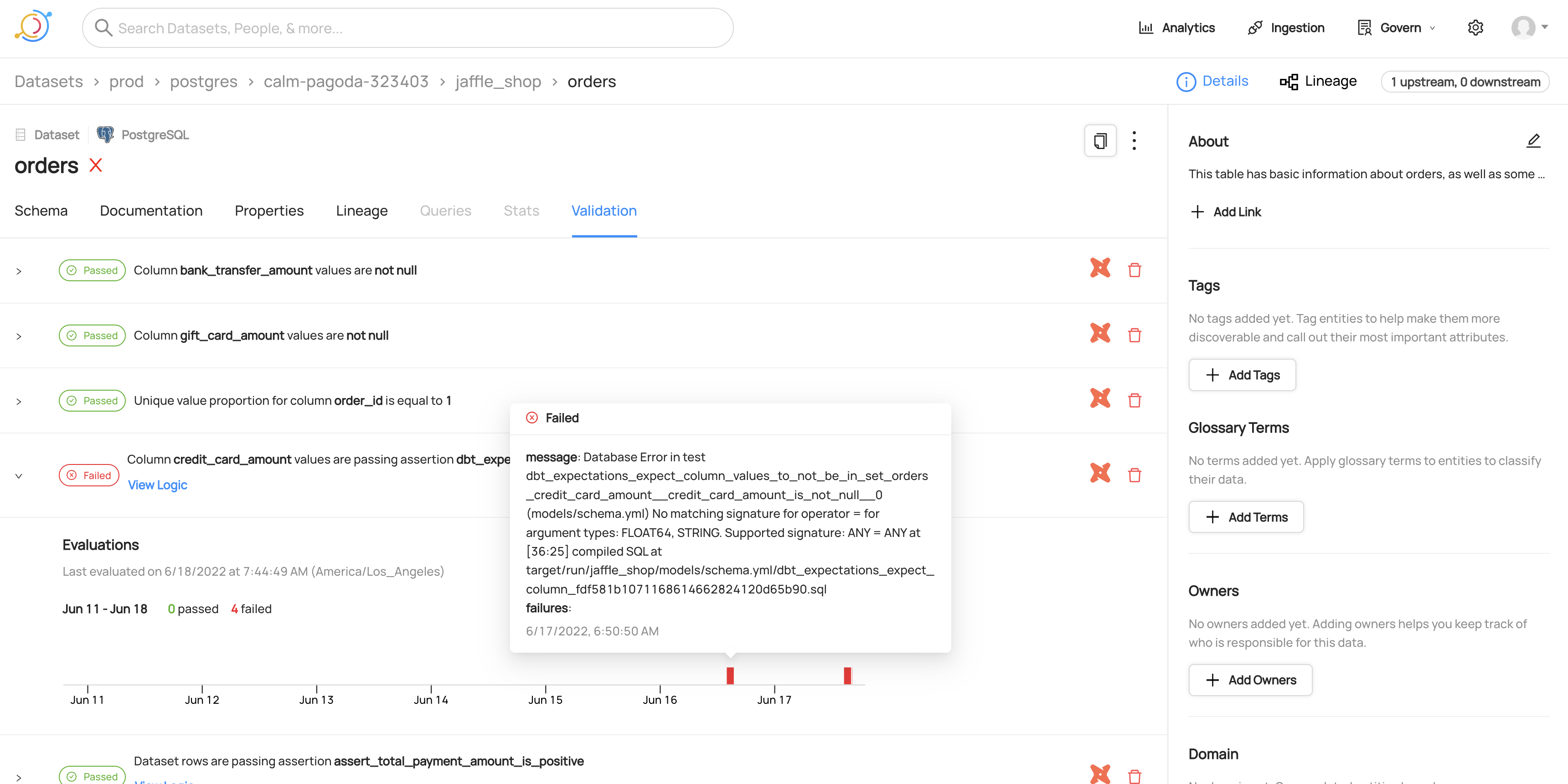
|
||||
|
||||
|
||||
|
||||
|
||||
@ -2,13 +2,14 @@ source:
|
||||
type: "dbt"
|
||||
config:
|
||||
# Coordinates
|
||||
manifest_path: "./path/dbt/manifest_file.json"
|
||||
catalog_path: "./path/dbt/catalog_file.json"
|
||||
sources_path: "./path/dbt/sources_file.json"
|
||||
# To use this as-is, set the environment variable DBT_PROJECT_ROOT to the root folder of your dbt project
|
||||
manifest_path: "${DBT_PROJECT_ROOT}/target/manifest_file.json"
|
||||
catalog_path: "${DBT_PROJECT_ROOT}/target/catalog_file.json"
|
||||
sources_path: "${DBT_PROJECT_ROOT}/target/sources_file.json" # optional for freshness
|
||||
test_results_path: "${DBT_PROJECT_ROOT}/target/run_results.json" # optional for recording dbt test results after running dbt test
|
||||
|
||||
# Options
|
||||
target_platform: "my_target_platform_id" # e.g. bigquery/postgres/etc.
|
||||
load_schemas: True # note: if this is disabled
|
||||
load_schemas: False # note: enable this only if you are not ingesting metadata from your warehouse
|
||||
|
||||
sink:
|
||||
# sink configs
|
||||
# sink configs
|
||||
|
||||
@ -2,12 +2,24 @@ import json
|
||||
import logging
|
||||
import re
|
||||
from dataclasses import dataclass, field
|
||||
from typing import Any, Dict, Iterable, List, Optional, Tuple, Type, cast
|
||||
from datetime import datetime
|
||||
from typing import (
|
||||
Any,
|
||||
Callable,
|
||||
Dict,
|
||||
Iterable,
|
||||
List,
|
||||
Optional,
|
||||
Tuple,
|
||||
Type,
|
||||
Union,
|
||||
cast,
|
||||
)
|
||||
from urllib.parse import urlparse
|
||||
|
||||
import dateutil.parser
|
||||
import requests
|
||||
from pydantic import validator
|
||||
from pydantic import BaseModel, validator
|
||||
from pydantic.fields import Field
|
||||
|
||||
from datahub.configuration.common import AllowDenyPattern, ConfigurationError
|
||||
@ -67,7 +79,21 @@ from datahub.metadata.com.linkedin.pegasus2avro.schema import (
|
||||
TimeTypeClass,
|
||||
)
|
||||
from datahub.metadata.schema_classes import (
|
||||
AssertionInfoClass,
|
||||
AssertionResultClass,
|
||||
AssertionResultTypeClass,
|
||||
AssertionRunEventClass,
|
||||
AssertionRunStatusClass,
|
||||
AssertionStdAggregationClass,
|
||||
AssertionStdOperatorClass,
|
||||
AssertionStdParameterClass,
|
||||
AssertionStdParametersClass,
|
||||
AssertionStdParameterTypeClass,
|
||||
AssertionTypeClass,
|
||||
ChangeTypeClass,
|
||||
DataPlatformInstanceClass,
|
||||
DatasetAssertionInfoClass,
|
||||
DatasetAssertionScopeClass,
|
||||
DatasetPropertiesClass,
|
||||
GlobalTagsClass,
|
||||
GlossaryTermsClass,
|
||||
@ -116,12 +142,16 @@ class DBTConfig(StatefulIngestionConfigBase):
|
||||
default=None,
|
||||
description="Path to dbt sources JSON. See https://docs.getdbt.com/reference/artifacts/sources-json. If not specified, last-modified fields will not be populated. Note this can be a local file or a URI.",
|
||||
)
|
||||
test_results_path: Optional[str] = Field(
|
||||
default=None,
|
||||
description="Path to output of dbt test run as run_results file in JSON format. See https://docs.getdbt.com/reference/artifacts/run-results-json. If not specified, test execution results will not be populated in DataHub.",
|
||||
)
|
||||
env: str = Field(
|
||||
default=mce_builder.DEFAULT_ENV,
|
||||
description="Environment to use in namespace when constructing URNs.",
|
||||
)
|
||||
target_platform: str = Field(
|
||||
description="The platform that dbt is loading onto. (e.g. bigquery / redshift / postgres etc.)"
|
||||
description="The platform that dbt is loading onto. (e.g. bigquery / redshift / postgres etc.)",
|
||||
)
|
||||
target_platform_instance: Optional[str] = Field(
|
||||
default=None,
|
||||
@ -182,6 +212,10 @@ class DBTConfig(StatefulIngestionConfigBase):
|
||||
default=None,
|
||||
description="When fetching manifest files from s3, configuration for aws connection details",
|
||||
)
|
||||
delete_tests_as_datasets: bool = Field(
|
||||
False,
|
||||
description="Prior to version 0.8.38, dbt tests were represented as datasets. If you ingested dbt tests before, set this flag to True (just needed once) to soft-delete tests that were generated as datasets by previous ingestion.",
|
||||
)
|
||||
|
||||
@property
|
||||
def s3_client(self):
|
||||
@ -271,6 +305,7 @@ class DBTNode:
|
||||
query_tag: Dict[str, Any] = field(default_factory=dict)
|
||||
|
||||
tags: List[str] = field(default_factory=list)
|
||||
compiled_sql: Optional[str] = None
|
||||
|
||||
def __repr__(self):
|
||||
fields = tuple("{}={}".format(k, v) for k, v in self.__dict__.items())
|
||||
@ -324,6 +359,7 @@ def extract_dbt_entities(
|
||||
continue
|
||||
|
||||
name = manifest_node["name"]
|
||||
|
||||
if "identifier" in manifest_node and use_identifiers:
|
||||
name = manifest_node["identifier"]
|
||||
|
||||
@ -355,10 +391,11 @@ def extract_dbt_entities(
|
||||
catalog_type = None
|
||||
|
||||
if catalog_node is None:
|
||||
report.report_warning(
|
||||
key,
|
||||
f"Entity {key} ({name}) is in manifest but missing from catalog",
|
||||
)
|
||||
if materialization != "test":
|
||||
report.report_warning(
|
||||
key,
|
||||
f"Entity {key} ({name}) is in manifest but missing from catalog",
|
||||
)
|
||||
else:
|
||||
catalog_type = all_catalog_entities[key]["metadata"]["type"]
|
||||
|
||||
@ -394,12 +431,14 @@ def extract_dbt_entities(
|
||||
query_tag=query_tag_props,
|
||||
tags=tags,
|
||||
owner=owner,
|
||||
compiled_sql=manifest_node.get("compiled_sql"),
|
||||
)
|
||||
|
||||
# overwrite columns from catalog
|
||||
if (
|
||||
dbtNode.materialization != "ephemeral"
|
||||
): # we don't want columns if platform isn't 'dbt'
|
||||
if dbtNode.materialization not in [
|
||||
"ephemeral",
|
||||
"test",
|
||||
]: # we don't want columns if platform isn't 'dbt'
|
||||
logger.debug("Loading schema info")
|
||||
catalog_node = all_catalog_entities.get(key)
|
||||
|
||||
@ -640,6 +679,231 @@ def get_schema_metadata(
|
||||
)
|
||||
|
||||
|
||||
@dataclass
|
||||
class AssertionParams:
|
||||
scope: Union[DatasetAssertionScopeClass, str]
|
||||
operator: Union[AssertionStdOperatorClass, str]
|
||||
aggregation: Union[AssertionStdAggregationClass, str]
|
||||
parameters: Optional[Callable[[Dict[str, str]], AssertionStdParametersClass]] = None
|
||||
logic_fn: Optional[Callable[[Dict[str, str]], Optional[str]]] = None
|
||||
|
||||
|
||||
def _get_name_for_relationship_test(kw_args: Dict[str, str]) -> Optional[str]:
|
||||
"""
|
||||
Try to produce a useful string for the name of a relationship constraint.
|
||||
Return None if we fail to
|
||||
"""
|
||||
destination_ref = kw_args.get("to")
|
||||
source_ref = kw_args.get("model")
|
||||
column_name = kw_args.get("column_name")
|
||||
dest_field_name = kw_args.get("field")
|
||||
if not destination_ref or not source_ref or not column_name or not dest_field_name:
|
||||
# base assertions are violated, bail early
|
||||
return None
|
||||
m = re.match(r"^ref\(\'(.*)\'\)$", destination_ref)
|
||||
if m:
|
||||
destination_table = m.group(1)
|
||||
else:
|
||||
destination_table = destination_ref
|
||||
m = re.search(r"ref\(\'(.*)\'\)", source_ref)
|
||||
if m:
|
||||
source_table = m.group(1)
|
||||
else:
|
||||
source_table = source_ref
|
||||
return f"{source_table}.{column_name} referential integrity to {destination_table}.{dest_field_name}"
|
||||
|
||||
|
||||
class DBTTestStep(BaseModel):
|
||||
name: Optional[str] = None
|
||||
started_at: Optional[str] = None
|
||||
completed_at: Optional[str] = None
|
||||
|
||||
|
||||
class DBTTestResult(BaseModel):
|
||||
class Config:
|
||||
extra = "allow"
|
||||
|
||||
status: str
|
||||
timing: List[DBTTestStep] = []
|
||||
unique_id: str
|
||||
failures: Optional[int] = None
|
||||
message: Optional[str] = None
|
||||
|
||||
|
||||
class DBTRunMetadata(BaseModel):
|
||||
dbt_schema_version: str
|
||||
dbt_version: str
|
||||
generated_at: str
|
||||
invocation_id: str
|
||||
|
||||
|
||||
class DBTTest:
|
||||
|
||||
test_name_to_assertion_map = {
|
||||
"not_null": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.NOT_NULL,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
),
|
||||
"unique": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.EQUAL_TO,
|
||||
aggregation=AssertionStdAggregationClass.UNIQUE_PROPOTION,
|
||||
parameters=lambda _: AssertionStdParametersClass(
|
||||
value=AssertionStdParameterClass(
|
||||
value="1.0",
|
||||
type=AssertionStdParameterTypeClass.NUMBER,
|
||||
)
|
||||
),
|
||||
),
|
||||
"accepted_values": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.IN,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
parameters=lambda kw_args: AssertionStdParametersClass(
|
||||
value=AssertionStdParameterClass(
|
||||
value=json.dumps(kw_args.get("values")),
|
||||
type=AssertionStdParameterTypeClass.SET,
|
||||
),
|
||||
),
|
||||
),
|
||||
"relationships": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass._NATIVE_,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
parameters=lambda kw_args: AssertionStdParametersClass(
|
||||
value=AssertionStdParameterClass(
|
||||
value=json.dumps(kw_args.get("values")),
|
||||
type=AssertionStdParameterTypeClass.SET,
|
||||
),
|
||||
),
|
||||
logic_fn=_get_name_for_relationship_test,
|
||||
),
|
||||
"dbt_expectations.expect_column_values_to_not_be_null": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.NOT_NULL,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
),
|
||||
"dbt_expectations.expect_column_values_to_be_between": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.BETWEEN,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
parameters=lambda x: AssertionStdParametersClass(
|
||||
minValue=AssertionStdParameterClass(
|
||||
value=str(x.get("min_value", "unknown")),
|
||||
type=AssertionStdParameterTypeClass.NUMBER,
|
||||
),
|
||||
maxValue=AssertionStdParameterClass(
|
||||
value=str(x.get("max_value", "unknown")),
|
||||
type=AssertionStdParameterTypeClass.NUMBER,
|
||||
),
|
||||
),
|
||||
),
|
||||
"dbt_expectations.expect_column_values_to_be_in_set": AssertionParams(
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass.IN,
|
||||
aggregation=AssertionStdAggregationClass.IDENTITY,
|
||||
parameters=lambda kw_args: AssertionStdParametersClass(
|
||||
value=AssertionStdParameterClass(
|
||||
value=json.dumps(kw_args.get("value_set")),
|
||||
type=AssertionStdParameterTypeClass.SET,
|
||||
),
|
||||
),
|
||||
),
|
||||
}
|
||||
|
||||
@staticmethod
|
||||
def load_test_results(
|
||||
config: DBTConfig,
|
||||
test_results_json: Dict[str, Any],
|
||||
test_nodes: List[DBTNode],
|
||||
manifest_nodes: Dict[str, Any],
|
||||
) -> Iterable[MetadataWorkUnit]:
|
||||
args = test_results_json.get("args", {})
|
||||
dbt_metadata = DBTRunMetadata.parse_obj(test_results_json.get("metadata", {}))
|
||||
test_nodes_map: Dict[str, DBTNode] = {x.dbt_name: x for x in test_nodes}
|
||||
if "test" in args.get("which", "") or "test" in args.get("rpc_method", ""):
|
||||
# this was a test run
|
||||
results = test_results_json.get("results", [])
|
||||
for result in results:
|
||||
try:
|
||||
test_result = DBTTestResult.parse_obj(result)
|
||||
id = test_result.unique_id
|
||||
test_node = test_nodes_map.get(id)
|
||||
assert test_node
|
||||
upstream_urns = get_upstreams(
|
||||
test_node.upstream_nodes,
|
||||
manifest_nodes,
|
||||
config.use_identifiers,
|
||||
config.target_platform,
|
||||
config.target_platform_instance,
|
||||
config.env,
|
||||
config.disable_dbt_node_creation,
|
||||
config.platform_instance,
|
||||
)
|
||||
assertion_urn = mce_builder.make_assertion_urn(
|
||||
mce_builder.datahub_guid(
|
||||
{
|
||||
"platform": DBT_PLATFORM,
|
||||
"name": test_result.unique_id,
|
||||
"instance": config.platform_instance,
|
||||
}
|
||||
)
|
||||
)
|
||||
|
||||
if test_result.status != "pass":
|
||||
native_results = {"message": test_result.message or ""}
|
||||
if test_result.failures:
|
||||
native_results.update(
|
||||
{"failures": str(test_result.failures)}
|
||||
)
|
||||
else:
|
||||
native_results = {}
|
||||
|
||||
stage_timings = {x.name: x.started_at for x in test_result.timing}
|
||||
# look for execution start time, fall back to compile start time and finally generation time
|
||||
execution_timestamp = (
|
||||
stage_timings.get("execute")
|
||||
or stage_timings.get("compile")
|
||||
or dbt_metadata.generated_at
|
||||
)
|
||||
|
||||
execution_timestamp_parsed = datetime.strptime(
|
||||
execution_timestamp, "%Y-%m-%dT%H:%M:%S.%fZ"
|
||||
)
|
||||
|
||||
for upstream in upstream_urns:
|
||||
assertionResult = AssertionRunEventClass(
|
||||
timestampMillis=int(
|
||||
execution_timestamp_parsed.timestamp() * 1000.0
|
||||
),
|
||||
assertionUrn=assertion_urn,
|
||||
asserteeUrn=upstream,
|
||||
runId=dbt_metadata.invocation_id,
|
||||
result=AssertionResultClass(
|
||||
type=AssertionResultTypeClass.SUCCESS
|
||||
if test_result.status == "pass"
|
||||
else AssertionResultTypeClass.FAILURE,
|
||||
nativeResults=native_results,
|
||||
),
|
||||
status=AssertionRunStatusClass.COMPLETE,
|
||||
)
|
||||
|
||||
event = MetadataChangeProposalWrapper(
|
||||
entityType="assertion",
|
||||
entityUrn=assertion_urn,
|
||||
changeType=ChangeTypeClass.UPSERT,
|
||||
aspectName="assertionRunEvent",
|
||||
aspect=assertionResult,
|
||||
)
|
||||
yield MetadataWorkUnit(
|
||||
id=f"{assertion_urn}-assertionRunEvent-{upstream}",
|
||||
mcp=event,
|
||||
)
|
||||
except Exception as e:
|
||||
logger.debug(f"Failed to process test result {result} due to {e}")
|
||||
|
||||
|
||||
@platform_name("dbt")
|
||||
@config_class(DBTConfig)
|
||||
@support_status(SupportStatus.CERTIFIED)
|
||||
@ -654,7 +918,7 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
- dbt Ephemeral: for nodes in the dbt manifest file that are ephemeral models
|
||||
- dbt Sources: for nodes that are sources on top of the underlying platform tables
|
||||
- dbt Seed: for seed entities
|
||||
- dbt Test: for dbt test entities
|
||||
- dbt Tests as Assertions: for dbt test entities (starting with version 0.8.38.1)
|
||||
|
||||
Note:
|
||||
1. It also generates lineage between the `dbt` nodes (e.g. ephemeral nodes that depend on other dbt sources) as well as lineage between the `dbt` nodes and the underlying (target) platform nodes (e.g. BigQuery Table -> dbt Source, dbt View -> BigQuery View).
|
||||
@ -663,7 +927,7 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
|
||||
The artifacts used by this source are:
|
||||
- [dbt manifest file](https://docs.getdbt.com/reference/artifacts/manifest-json)
|
||||
- This file contains model, source and lineage data.
|
||||
- This file contains model, source, tests and lineage data.
|
||||
- [dbt catalog file](https://docs.getdbt.com/reference/artifacts/catalog-json)
|
||||
- This file contains schema data.
|
||||
- dbt does not record schema data for Ephemeral models, as such datahub will show Ephemeral models in the lineage, however there will be no associated schema for Ephemeral models
|
||||
@ -671,7 +935,9 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
- This file contains metadata for sources with freshness checks.
|
||||
- We transfer dbt's freshness checks to DataHub's last-modified fields.
|
||||
- Note that this file is optional – if not specified, we'll use time of ingestion instead as a proxy for time last-modified.
|
||||
|
||||
- [dbt run_results file](https://docs.getdbt.com/reference/artifacts/run-results-json)
|
||||
- This file contains metadata from the result of a dbt run, e.g. dbt test
|
||||
- When provided, we transfer dbt test run results into assertion run events to see a timeline of test runs on the dataset
|
||||
"""
|
||||
|
||||
@classmethod
|
||||
@ -736,7 +1002,6 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
):
|
||||
yield from soft_delete_item(table_urn, "dataset")
|
||||
|
||||
# s3://data-analysis.pelotime.com/dbt-artifacts/data-engineering-dbt/catalog.json
|
||||
def load_file_as_json(self, uri: str) -> Any:
|
||||
if re.match("^https?://", uri):
|
||||
return json.loads(requests.get(uri).text)
|
||||
@ -818,6 +1083,193 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
all_manifest_entities,
|
||||
)
|
||||
|
||||
def create_test_entity_mcps(
|
||||
self,
|
||||
test_nodes: List[DBTNode],
|
||||
custom_props: Dict[str, str],
|
||||
manifest_nodes: Dict[str, Dict[str, Any]],
|
||||
) -> Iterable[MetadataWorkUnit]:
|
||||
def string_map(input_map: Dict[str, Any]) -> Dict[str, str]:
|
||||
return {k: str(v) for k, v in input_map.items()}
|
||||
|
||||
if self.config.test_results_path:
|
||||
yield from DBTTest.load_test_results(
|
||||
self.config,
|
||||
self.load_file_as_json(self.config.test_results_path),
|
||||
test_nodes,
|
||||
manifest_nodes,
|
||||
)
|
||||
|
||||
for node in test_nodes:
|
||||
node_datahub_urn = mce_builder.make_assertion_urn(
|
||||
mce_builder.datahub_guid(
|
||||
{
|
||||
"platform": DBT_PLATFORM,
|
||||
"name": node.dbt_name,
|
||||
"instance": self.config.platform_instance,
|
||||
}
|
||||
)
|
||||
)
|
||||
self.save_checkpoint(node_datahub_urn)
|
||||
|
||||
dpi_mcp = MetadataChangeProposalWrapper(
|
||||
entityType="assertion",
|
||||
entityUrn=node_datahub_urn,
|
||||
changeType=ChangeTypeClass.UPSERT,
|
||||
aspectName="dataPlatformInstance",
|
||||
aspect=DataPlatformInstanceClass(
|
||||
platform=mce_builder.make_data_platform_urn(DBT_PLATFORM)
|
||||
),
|
||||
)
|
||||
wu = MetadataWorkUnit(
|
||||
id=f"{node_datahub_urn}-dataplatformInstance", mcp=dpi_mcp
|
||||
)
|
||||
self.report.report_workunit(wu)
|
||||
yield wu
|
||||
|
||||
upstream_urns = get_upstreams(
|
||||
upstreams=node.upstream_nodes,
|
||||
all_nodes=manifest_nodes,
|
||||
use_identifiers=self.config.use_identifiers,
|
||||
target_platform=self.config.target_platform,
|
||||
target_platform_instance=self.config.target_platform_instance,
|
||||
environment=self.config.env,
|
||||
disable_dbt_node_creation=self.config.disable_dbt_node_creation,
|
||||
platform_instance=None,
|
||||
)
|
||||
|
||||
raw_node = manifest_nodes.get(node.dbt_name)
|
||||
if raw_node is None:
|
||||
logger.warning(
|
||||
f"Failed to find test node {node.dbt_name} in the manifest"
|
||||
)
|
||||
continue
|
||||
|
||||
test_metadata = raw_node.get("test_metadata", {})
|
||||
kw_args = test_metadata.get("kwargs", {})
|
||||
for upstream_urn in upstream_urns:
|
||||
qualified_test_name = (
|
||||
(test_metadata.get("namespace") or "")
|
||||
+ "."
|
||||
+ (test_metadata.get("name") or "")
|
||||
)
|
||||
qualified_test_name = (
|
||||
qualified_test_name[1:]
|
||||
if qualified_test_name.startswith(".")
|
||||
else qualified_test_name
|
||||
)
|
||||
|
||||
if qualified_test_name in DBTTest.test_name_to_assertion_map:
|
||||
assertion_params: AssertionParams = (
|
||||
DBTTest.test_name_to_assertion_map[qualified_test_name]
|
||||
)
|
||||
assertion_info = AssertionInfoClass(
|
||||
type=AssertionTypeClass.DATASET,
|
||||
customProperties=custom_props,
|
||||
datasetAssertion=DatasetAssertionInfoClass(
|
||||
dataset=upstream_urn,
|
||||
scope=assertion_params.scope,
|
||||
operator=assertion_params.operator,
|
||||
fields=[
|
||||
mce_builder.make_schema_field_urn(
|
||||
upstream_urn, kw_args.get("column_name")
|
||||
)
|
||||
]
|
||||
if assertion_params.scope
|
||||
== DatasetAssertionScopeClass.DATASET_COLUMN
|
||||
else [],
|
||||
nativeType=node.name,
|
||||
aggregation=assertion_params.aggregation,
|
||||
parameters=assertion_params.parameters(kw_args)
|
||||
if assertion_params.parameters
|
||||
else None,
|
||||
logic=assertion_params.logic_fn(kw_args)
|
||||
if assertion_params.logic_fn
|
||||
else None,
|
||||
nativeParameters=string_map(kw_args),
|
||||
),
|
||||
)
|
||||
elif kw_args.get("column_name"):
|
||||
# no match with known test types, column-level test
|
||||
assertion_info = AssertionInfoClass(
|
||||
type=AssertionTypeClass.DATASET,
|
||||
customProperties=custom_props,
|
||||
datasetAssertion=DatasetAssertionInfoClass(
|
||||
dataset=upstream_urn,
|
||||
scope=DatasetAssertionScopeClass.DATASET_COLUMN,
|
||||
operator=AssertionStdOperatorClass._NATIVE_,
|
||||
fields=[
|
||||
mce_builder.make_schema_field_urn(
|
||||
upstream_urn, kw_args.get("column_name")
|
||||
)
|
||||
],
|
||||
nativeType=node.name,
|
||||
logic=node.compiled_sql
|
||||
if node.compiled_sql
|
||||
else node.raw_sql,
|
||||
aggregation=AssertionStdAggregationClass._NATIVE_,
|
||||
nativeParameters=string_map(kw_args),
|
||||
),
|
||||
)
|
||||
else:
|
||||
# no match with known test types, default to row-level test
|
||||
assertion_info = AssertionInfoClass(
|
||||
type=AssertionTypeClass.DATASET,
|
||||
customProperties=custom_props,
|
||||
datasetAssertion=DatasetAssertionInfoClass(
|
||||
dataset=upstream_urn,
|
||||
scope=DatasetAssertionScopeClass.DATASET_ROWS,
|
||||
operator=AssertionStdOperatorClass._NATIVE_,
|
||||
logic=node.compiled_sql
|
||||
if node.compiled_sql
|
||||
else node.raw_sql,
|
||||
nativeType=node.name,
|
||||
aggregation=AssertionStdAggregationClass._NATIVE_,
|
||||
nativeParameters=string_map(kw_args),
|
||||
),
|
||||
)
|
||||
wu = MetadataWorkUnit(
|
||||
id=f"{node_datahub_urn}-assertioninfo",
|
||||
mcp=MetadataChangeProposalWrapper(
|
||||
entityType="assertion",
|
||||
entityUrn=node_datahub_urn,
|
||||
changeType=ChangeTypeClass.UPSERT,
|
||||
aspectName="assertionInfo",
|
||||
aspect=assertion_info,
|
||||
),
|
||||
)
|
||||
self.report.report_workunit(wu)
|
||||
yield wu
|
||||
|
||||
if self.config.delete_tests_as_datasets:
|
||||
mce_platform = (
|
||||
self.config.target_platform
|
||||
if self.config.disable_dbt_node_creation
|
||||
else DBT_PLATFORM
|
||||
)
|
||||
node_datahub_urn = get_urn_from_dbtNode(
|
||||
node.database,
|
||||
node.schema,
|
||||
node.name,
|
||||
mce_platform,
|
||||
self.config.env,
|
||||
self.config.platform_instance
|
||||
if mce_platform == DBT_PLATFORM
|
||||
else None,
|
||||
)
|
||||
soft_delete_mcp = MetadataChangeProposalWrapper(
|
||||
entityType="dataset",
|
||||
changeType=ChangeTypeClass.UPSERT,
|
||||
entityUrn=node_datahub_urn,
|
||||
aspectName="status",
|
||||
aspect=StatusClass(removed=True),
|
||||
)
|
||||
soft_delete_wu = MetadataWorkUnit(
|
||||
id=f"{node_datahub_urn}-status", mcp=soft_delete_mcp
|
||||
)
|
||||
self.report.report_workunit(soft_delete_wu)
|
||||
yield soft_delete_wu
|
||||
|
||||
# create workunits from dbt nodes
|
||||
def get_workunits(self) -> Iterable[MetadataWorkUnit]:
|
||||
if self.config.write_semantics == "PATCH" and not self.ctx.graph:
|
||||
@ -858,9 +1310,14 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
if value is not None
|
||||
}
|
||||
|
||||
non_test_nodes = [
|
||||
dataset_node for dataset_node in nodes if dataset_node.node_type != "test"
|
||||
]
|
||||
test_nodes = [test_node for test_node in nodes if test_node.node_type == "test"]
|
||||
|
||||
if not self.config.disable_dbt_node_creation:
|
||||
yield from self.create_platform_mces(
|
||||
nodes,
|
||||
non_test_nodes,
|
||||
additional_custom_props_filtered,
|
||||
manifest_nodes_raw,
|
||||
DBT_PLATFORM,
|
||||
@ -868,13 +1325,19 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
)
|
||||
|
||||
yield from self.create_platform_mces(
|
||||
nodes,
|
||||
non_test_nodes,
|
||||
additional_custom_props_filtered,
|
||||
manifest_nodes_raw,
|
||||
self.config.target_platform,
|
||||
self.config.target_platform_instance,
|
||||
)
|
||||
|
||||
yield from self.create_test_entity_mcps(
|
||||
test_nodes,
|
||||
additional_custom_props_filtered,
|
||||
manifest_nodes_raw,
|
||||
)
|
||||
|
||||
if self.is_stateful_ingestion_configured():
|
||||
# Clean up stale entities.
|
||||
yield from self.gen_removed_entity_workunits()
|
||||
@ -1050,25 +1513,6 @@ class DBTSource(StatefulIngestionSourceBase):
|
||||
return aspect
|
||||
return None
|
||||
|
||||
# TODO: Remove. keeping this till PR review
|
||||
# def get_owners_from_dataset_snapshot(self, dataset_snapshot: DatasetSnapshot) -> Optional[OwnershipClass]:
|
||||
# for aspect in dataset_snapshot.aspects:
|
||||
# if isinstance(aspect, OwnershipClass):
|
||||
# return aspect
|
||||
# return None
|
||||
#
|
||||
# def get_tag_aspect_from_dataset_snapshot(self, dataset_snapshot: DatasetSnapshot) -> Optional[GlobalTagsClass]:
|
||||
# for aspect in dataset_snapshot.aspects:
|
||||
# if isinstance(aspect, GlobalTagsClass):
|
||||
# return aspect
|
||||
# return None
|
||||
#
|
||||
# def get_term_aspect_from_dataset_snapshot(self, dataset_snapshot: DatasetSnapshot) -> Optional[GlossaryTermsClass]:
|
||||
# for aspect in dataset_snapshot.aspects:
|
||||
# if isinstance(aspect, GlossaryTermsClass):
|
||||
# return aspect
|
||||
# return None
|
||||
|
||||
def get_patched_mce(self, mce):
|
||||
owner_aspect = self.get_aspect_from_dataset(
|
||||
mce.proposedSnapshot, OwnershipClass
|
||||
|
||||
3763
metadata-ingestion/tests/integration/dbt/dbt_test_events_golden.json
Normal file
3763
metadata-ingestion/tests/integration/dbt/dbt_test_events_golden.json
Normal file
File diff suppressed because it is too large
Load Diff
File diff suppressed because one or more lines are too long
File diff suppressed because one or more lines are too long
File diff suppressed because one or more lines are too long
@ -6,8 +6,9 @@ import pytest
|
||||
import requests_mock
|
||||
from freezegun import freeze_time
|
||||
|
||||
from datahub.ingestion.run.pipeline import Pipeline
|
||||
from datahub.ingestion.source.dbt import DBTSource
|
||||
from datahub.configuration.common import DynamicTypedConfig
|
||||
from datahub.ingestion.run.pipeline import Pipeline, PipelineConfig, SourceConfig
|
||||
from datahub.ingestion.source.dbt import DBTConfig, DBTSource
|
||||
from datahub.ingestion.source.state.checkpoint import Checkpoint
|
||||
from datahub.ingestion.source.state.sql_common_state import (
|
||||
BaseSQLAlchemyCheckpointState,
|
||||
@ -421,3 +422,46 @@ def test_dbt_stateful(pytestconfig, tmp_path, mock_time, mock_datahub_graph):
|
||||
output_path=deleted_mces_path,
|
||||
golden_path=deleted_actor_golden_mcs,
|
||||
)
|
||||
|
||||
|
||||
@pytest.mark.integration
|
||||
@freeze_time(FROZEN_TIME)
|
||||
def test_dbt_tests(pytestconfig, tmp_path, mock_time, **kwargs):
|
||||
test_resources_dir = pytestconfig.rootpath / "tests/integration/dbt"
|
||||
|
||||
# Run the metadata ingestion pipeline.
|
||||
output_file = tmp_path / "dbt_test_events.json"
|
||||
golden_path = test_resources_dir / "dbt_test_events_golden.json"
|
||||
|
||||
pipeline = Pipeline(
|
||||
config=PipelineConfig(
|
||||
source=SourceConfig(
|
||||
type="dbt",
|
||||
config=DBTConfig(
|
||||
manifest_path=str(
|
||||
(test_resources_dir / "jaffle_shop_manifest.json").resolve()
|
||||
),
|
||||
catalog_path=str(
|
||||
(test_resources_dir / "jaffle_shop_catalog.json").resolve()
|
||||
),
|
||||
target_platform="postgres",
|
||||
delete_tests_as_datasets=True,
|
||||
test_results_path=str(
|
||||
(test_resources_dir / "jaffle_shop_test_results.json").resolve()
|
||||
),
|
||||
# this is just here to avoid needing to access datahub server
|
||||
write_semantics="OVERRIDE",
|
||||
),
|
||||
),
|
||||
sink=DynamicTypedConfig(type="file", config={"filename": str(output_file)}),
|
||||
)
|
||||
)
|
||||
pipeline.run()
|
||||
pipeline.raise_from_status()
|
||||
# Verify the output.
|
||||
mce_helpers.check_golden_file(
|
||||
pytestconfig,
|
||||
output_path=output_file,
|
||||
golden_path=golden_path,
|
||||
ignore_paths=[],
|
||||
)
|
||||
|
||||
Loading…
x
Reference in New Issue
Block a user